T1i / 14A10: Classify bacteria according to Gram stain system and their shape. Give two examples for each classification (40% marks). List with examples the mechanisms of bacterial antibiotic resistance (60% marks)
14A10: Exam Report
Classify bacteria according to the Gram stain system and their shape. Give two examples for each classification. (40% of marks) List with examples the mechanisms of bacterial antibiotic resistance. (60% of marks)
63% of candidates passed this question.
Generally candidates provided an accurate classification of bacteria based upon Gram staining and shape. None or poorly staining organisms were often overlooked. Mechanism of resistance was also well covered, with examples. Areas of weakness were a lack of descriptive detail and/or omission of an example for each mechanism. Mechanisms such as metabolic blockade of essential pathways for antibacterial action and active removal of intracellular antibiotic were more commonly omitted.
T1i / 14A10: Classify bacteria according to Gram stain system and their shape. Give two examples for each classification (40% marks). List with examples the mechanisms of bacterial antibiotic resistance (60% marks).
Definitions
Bacteria = Prokaryote – single cell organism lacking a nucleus or any membrane-bound organelles
Gram Stain
- Bacterial slides are first stained w Crystal Violet and Iodine
- Then destained w alcohol
- Then counter-stained w safranin
Gram +ve
- Blue purple
- Large peptidoglycan wall
- Crystal violet precipitates in wall and not affected by alcohol wash
Gram –ve
- Pink
- From the safranin stain
- Small peptidoglycan wall with w additional outer PM layer
Gram Indeterminate
- Bacteria which do not respond predictably to Gram staining or have a variable pink/purple pattern
- Cannot be classified as G +/-
- Myocabacterium, Clostridium
Morphology & Growth Requirements
Gram -ve
Anaerobic
Rods
Baceteriods
Cocci
Veillonella
Aerobic
Rods
Pseudomonas
E Coli
Klebsiella
Hemophilus
Cocci
Neisseria
Gram +ve
Anaerobic
Rods
Clostridium
Cocci
Staphylococci
Streptococci
Enterococci
Aerobic
Rods
Listeria
Cocci
Peptococcus
Additional Classification
Catalase
- Hydrogen peroxide added to bacterial sample
- Presence of catalase produced oxygen
- Catalase+ = Staphylococci
- Catalase- = Streptococci & Enterococci
Coagulase
- Performed on Staphylococcals
- Coagulase is an enzyme wh cleaves fibrinogen → fibrin
- Rabbit plasma added to bacterial sample
- If coagulase is present, fibrin will form
- Coag+ = S Aureus
- Coag- = S Epidermidis
Hemolytic
- Performed on Streptococcal
- Streptococcal colonies are added to blood agar
- Colour change = hemolysis
- α haemolytic
- dark green agar
- due to metHb produced by organism
- Strep pneumonia
- β haemolytic
- yellow agar
- complete hemolysis
- Strep pyogenes
- ϒ haemolytic
- Agar unchanged
- E faecalis
- E faecium
- α haemolytic
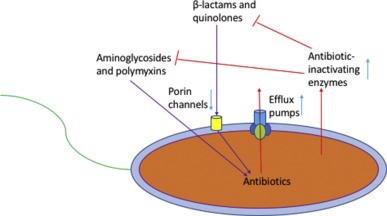
Resistance = when the maximal level of the agent tolerated is insufficient to inhibit growth
- Natural – bacteria do not possess molecular target for that drug
- Acquired:
1. Modification of antimicrobial molecule
a. Chemical Alteration of the Antibiotic
- Production of enzymes capable of introducing chemical changes to the antimicrobial molecule
- Example: Aminoglycoside Modifying Enzymes (AMEs) which modify the hydroxyl/amino groups of the aminoglycoside, enabling resistance E Faecium
b. Destruction of the Antibiotic Molecule
- b-lactamases produced by bacteria hydrolyse the b-lactam ring
- \negates penicillin
- still sensitive to cephalosporins
- ESBLs = even more widespread resistance
- R to cephalosporins/penicillin/b-lactamase inhibitors (Clav Acid)
- Only Mero is effective
2. Prevention of antibiotic reaching target
a. Decreased permeability
- Many a/b have targets inside bacteria or on their membrane
- Example: Vancomycin is not active against Gm- bacteria as it cannot penetrate the outer membrane
- Example: P aeruginosa is very good at altering porin fn/expression to limit permeability of a/b
b. Efflux pumps
- P aeruginosa possess an efflux system to pump a/b out of the cell
3. Change/Bypass of target site
- Changing of target site = ¯affinity of drug
- Example: VRE
- Glycopeptides bind to D-Ala-D-Ala
- Bacterial remodelling of cell wall to provide new substrate D-Ala-D-Lactate prevents Vancomycin binding
- Especially common in enterococci (E Faecium)
4. Resistance due to global cell adaptive processes
- Example: Daptomycin resistance
- Altered cell wall metabolism resulting in changes in surface charge of the bacteria and produces an electrostatic ‘repulsion’ of Daptomycin from the cell wall of enterococci
Pseudomonas Aeruginosa has been shown to possess a high level of intrinsic resistance to most a/b via
- Restricted outer membrane permeability
- Efflux systems
- Production of inactivating enzyme like b-lactamases
Methods of Resistance Spread
1. Transfer of bacteria between people
2. Transfer of R genes between bacteria
a. Conjugation – bacteria connect through tubing & transmit plasmid
b. Transduction – plasmid enclosed in ‘phage’ and transferred to another bacterium
c. Transformation – bac take up DNA from enviro and incorporate into their genome
- Author: Krisoula Zahariou